Navigation
What is dSCOPE
dSCOPE is a web server developed for detecting sequences critical for phase separation related proteins. Based on random forest algorithm, the predictor achieved promising performances with three bright spots as follows :ⅰ) Multiple features are extracted to build the prediction model; ⅱ) Various sequence and structural properties including phase separation region, disorder, exposed & buried, polar, low complexity region, charge, secondary structure, prion-like region, surface accessibility, hydropathy, subcellular location are visualized; ⅲ) Pre-predicted key regions of phase separation for human proteome are provided.
About predicting performance
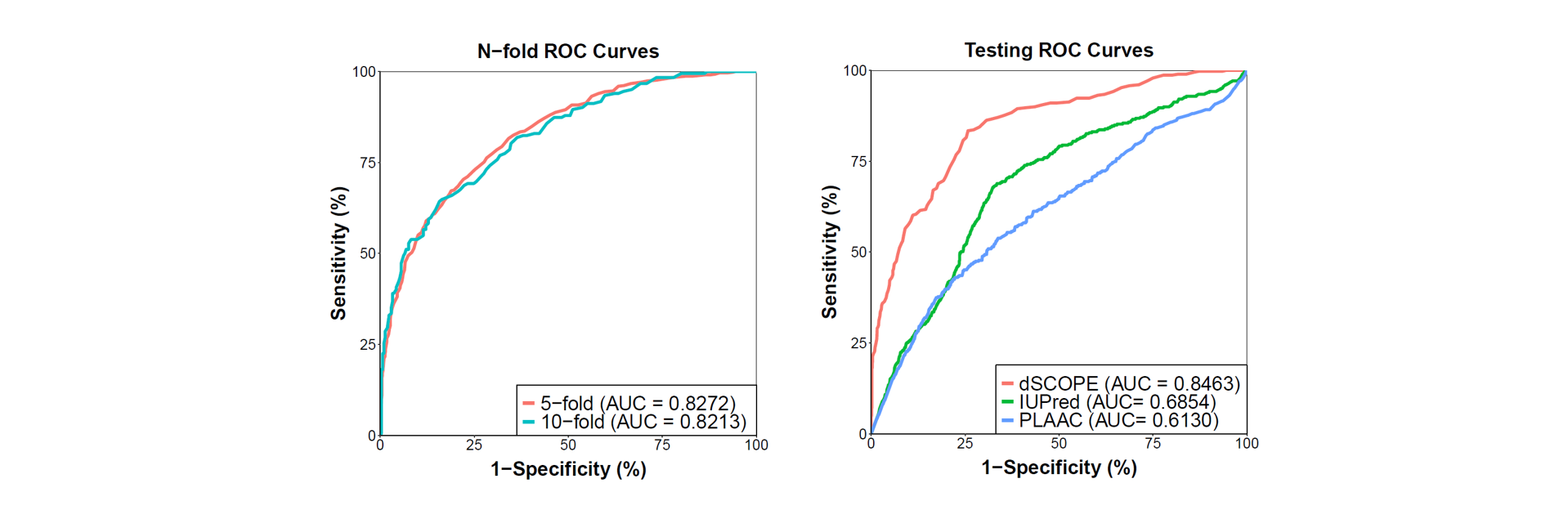
ROC for cross validation (left), ROC for dSCOPE compared to other features, including IUPred and PLAAC(right).
Usage of web server
Web server procedure
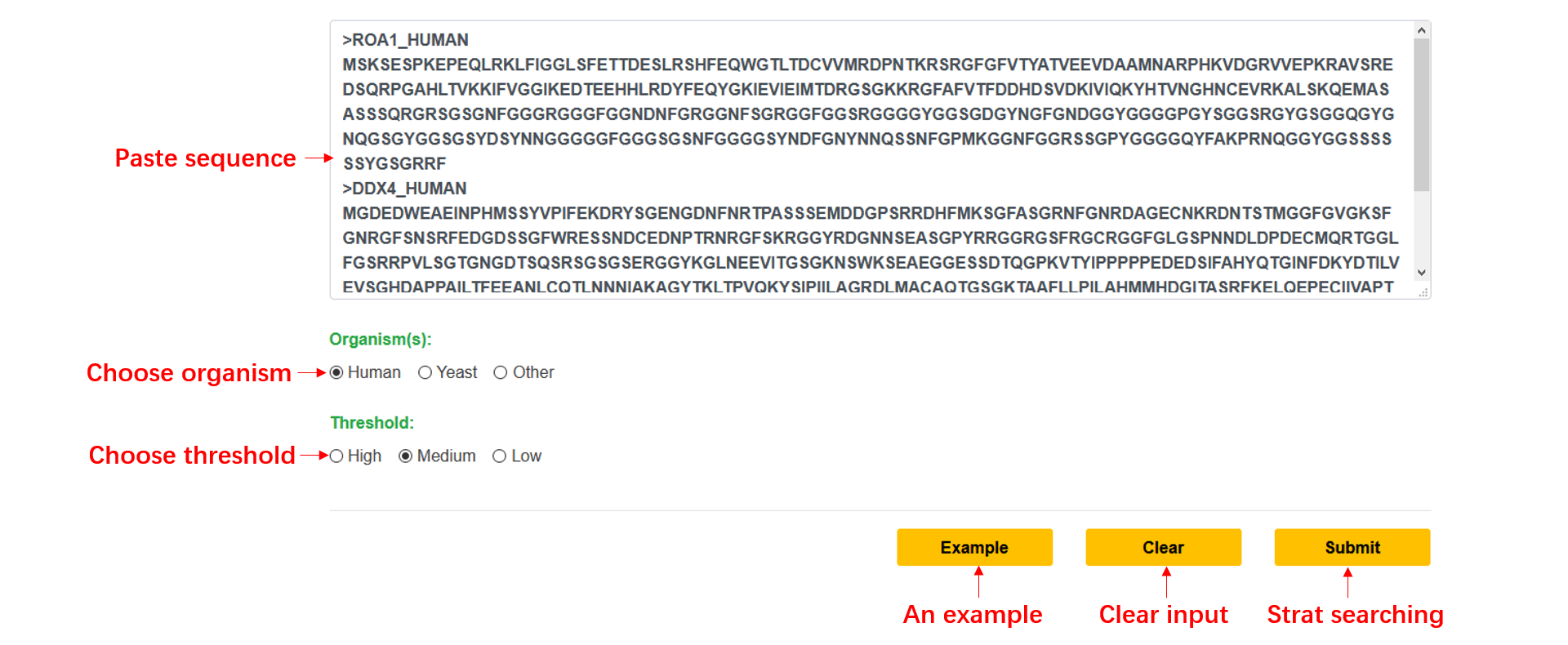
Step1: Paste your FASTA format sequence into the input textarea, or you can click the example button to run the default sequence.
Step2: Choose a kind of organism, along with a threshold, and click submit.
Web server result
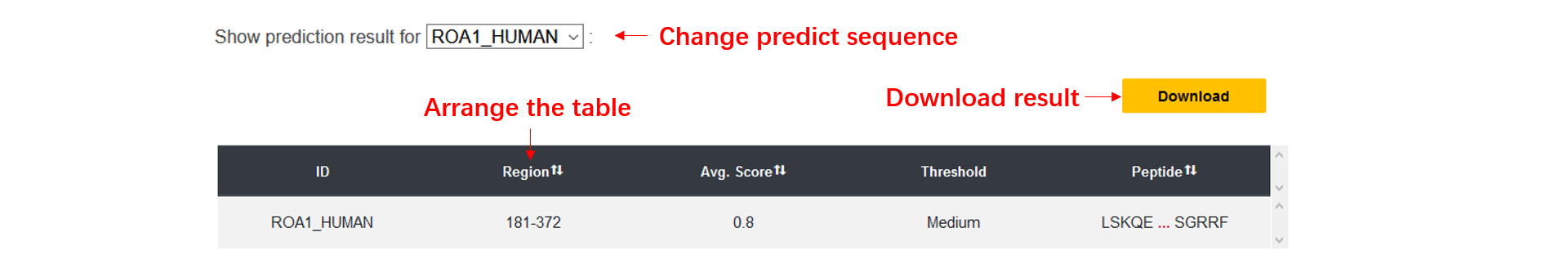
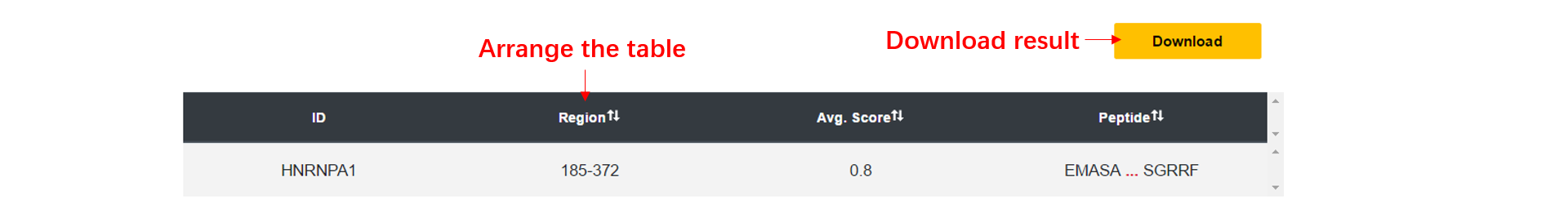
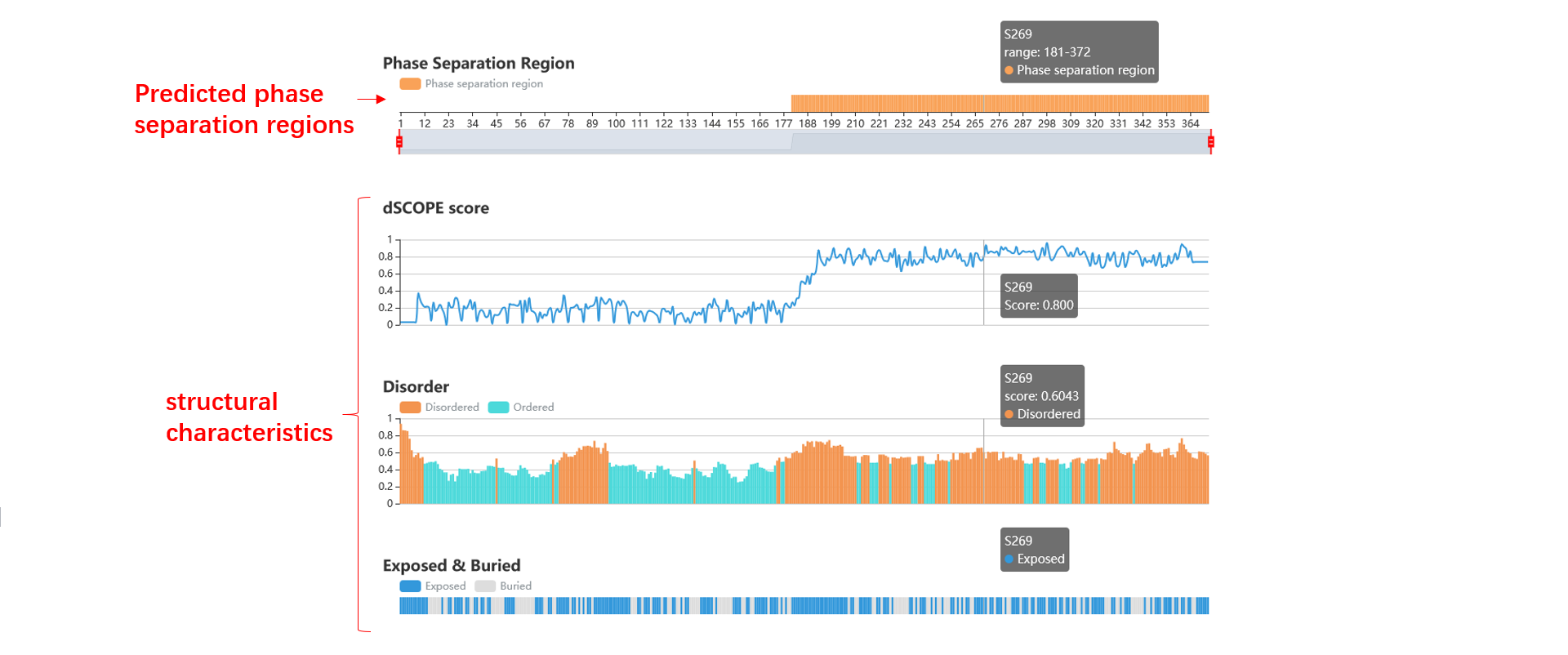
1. The dSCOPE prediction results including the phase separation region, the average dSCOPE score, the threshold and the peptide.
2. you can arrange the result table, and we only display the first five and the last five amino acids of the peptide, you can see the full sequence if you click it.
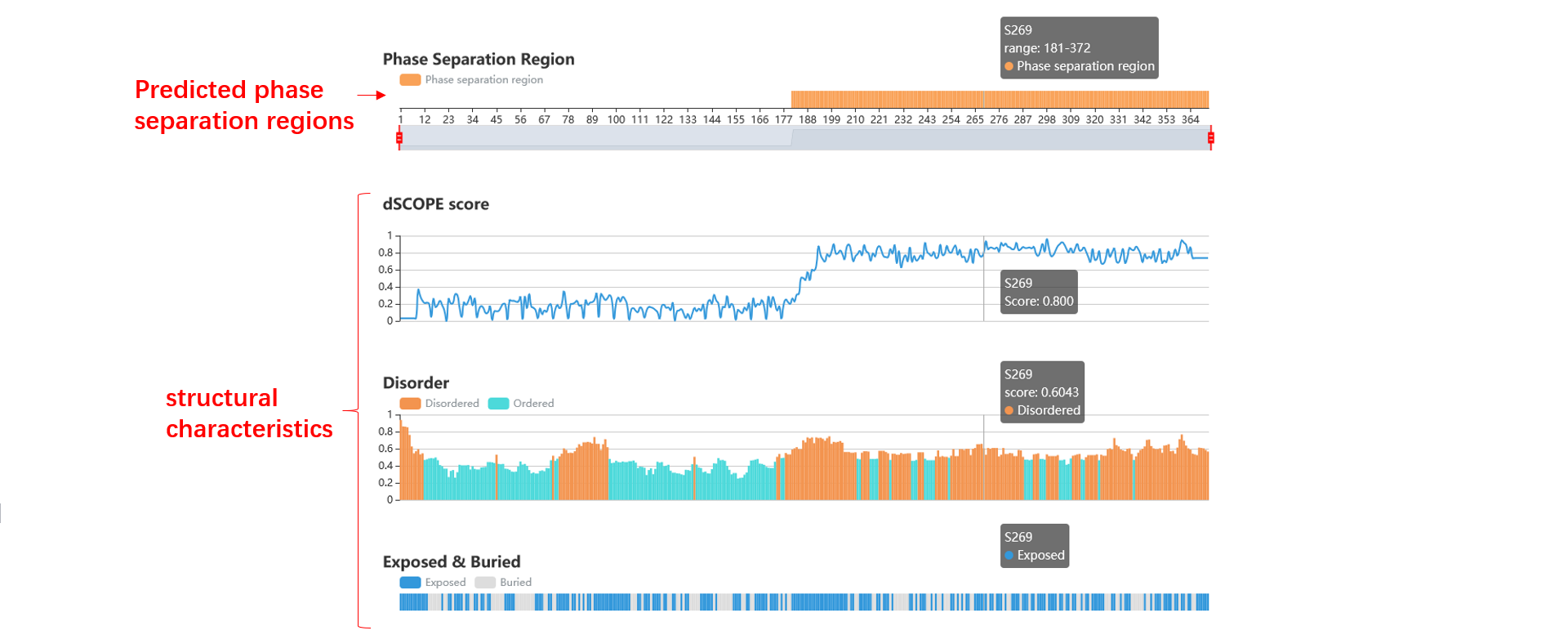
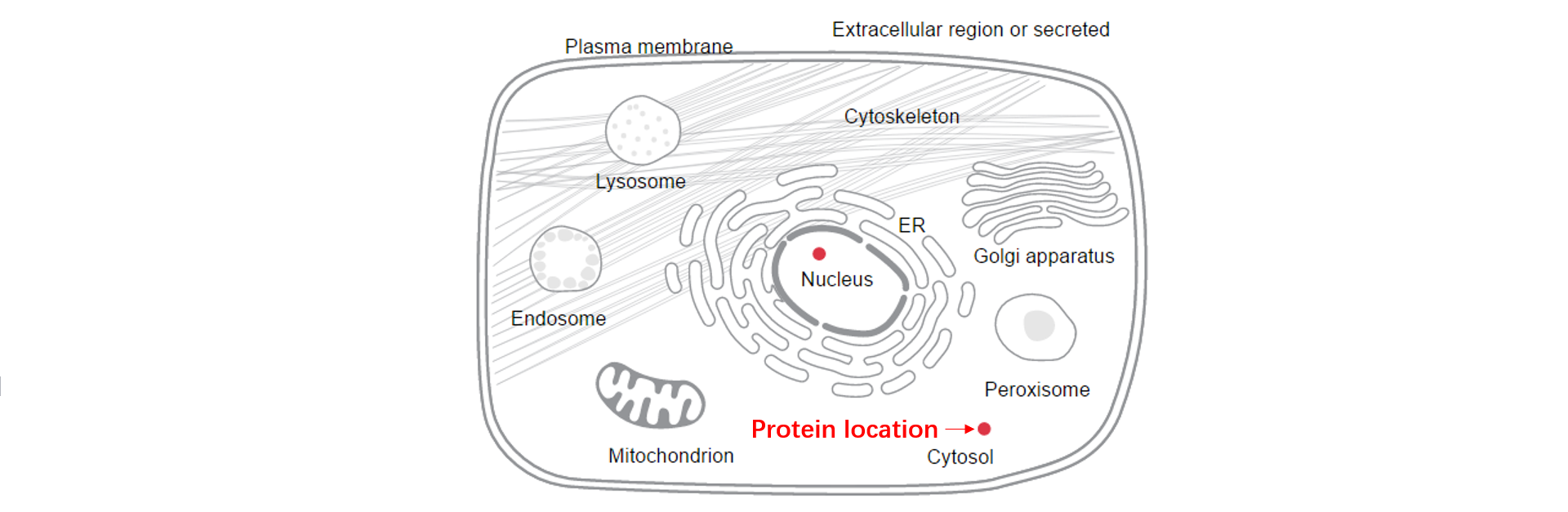
3. We dispaly the predicted phase separation region, the dSCOPE score of every amino acid, the structural characteristics (disorder, exposed or buried, polar, low complex, charge, second structure, prion-like, surface accessibility and hydropathy), and the subcellular location.
4. You can scale all the graphics by control element in the top, or through the mouse wheel, the detailed information will be shown if you hover the mouse on an amino acid.
Usage of database search
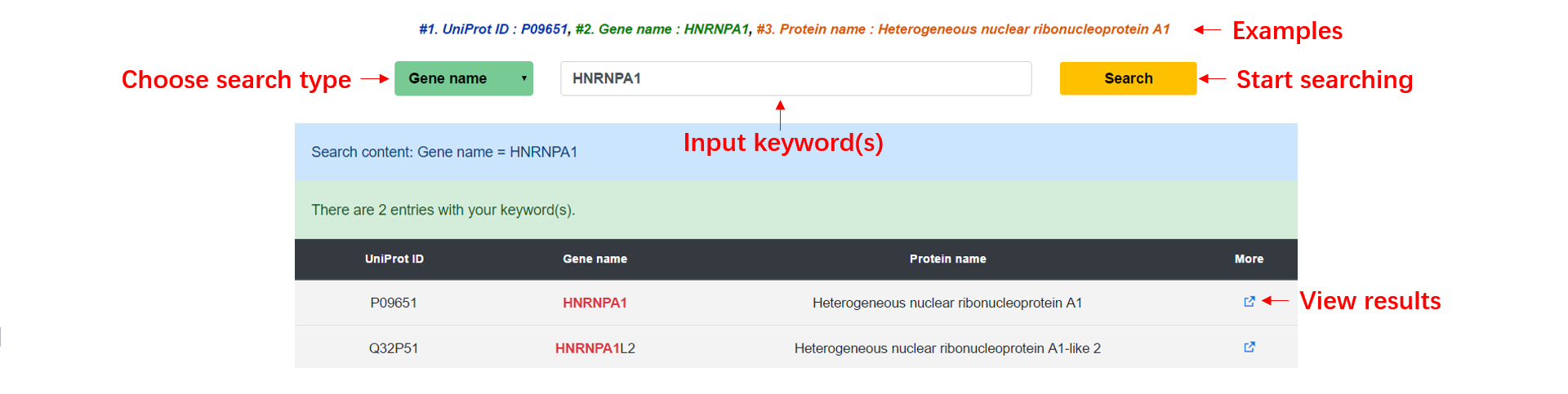
Database search procedure
Step1: You can choose a search type and enter a keyword to start searching the dSCOPE, and we provide an example for each search type, including UniProt ID, gene name and protein name.
Step2: All matched results will be displayed in a table at the bottom of the search page, you can click the more button to see the predicted result of the protein.
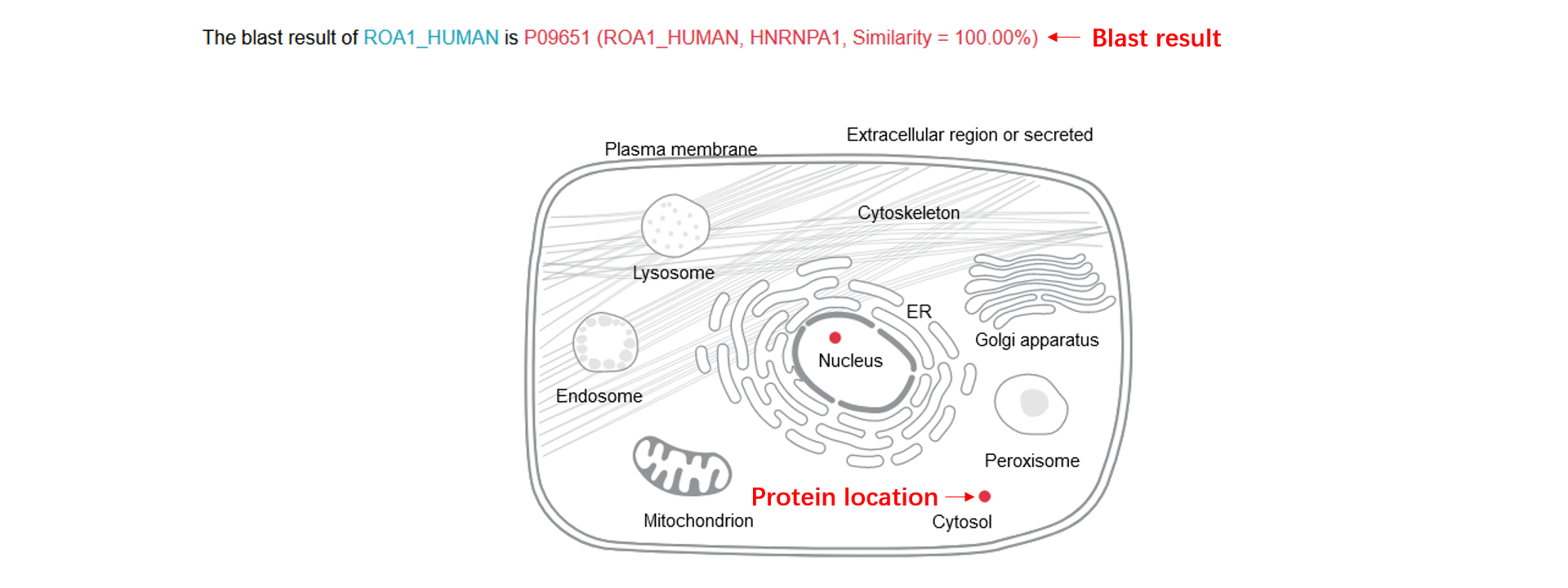
Database search result
1. The dSCOPE prediction results including the phase separation region, the average dSCOPE score, the threshold and the peptide.
2. you can arrange the result table, and we only display the first five and the last five amino acids of the peptide, you can see the full sequence if you click it.
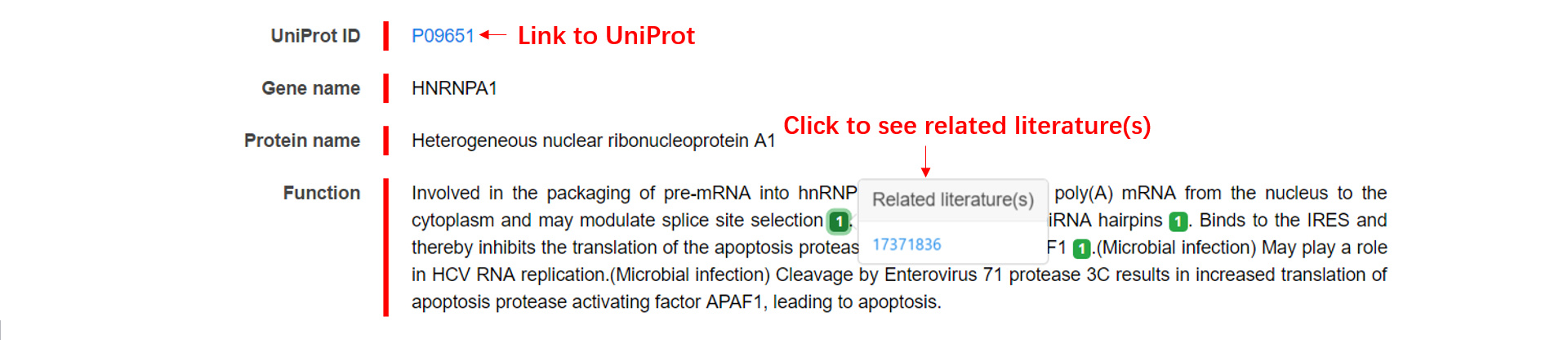
3. We collected the information of the protein from UniProt, and a corresponding link is provided.
4. We dispaly the predicted phase separation region, the dSCOPE score of every amino acid, the structural characteristics (disorder, exposed or buried, polar, low complex, charge, second structure, prion-like, surface accessibility and hydropathy), and the subcellular location.
5. You can scale all the graphics by control element in the top, or through the mouse wheel, the detailed information will be shown if you hover the mouse on an amino acid.
The implemented algorithm
To predict the sequence region that is important to phase separation, the random forest algorithm is introduced into our prediction. For a given protein sequence, a short peptide consisting of 15 amino acids in length is intercepted through a sliding window. Each adjacent short peptide needs to be separated by 8 amino acid residues, and short peptides less than 15 in length are removed. Protein fragments are encoded by eight physical and chemical features scores and four feature extraction methods. Next, we tested the performance of five ML classifiers, including logistic regression, random forest, LDA, AdaBoost, and KNN, then adopted the final algorithm for prediction based on the performance. Moreover, to generate the optimal performance, the TPOT (https://github.com/EpistasisLab/tpot) package was integrated to optimize the hyperparameters.
Methods for feature extraction
Physicochemical properties
In recent years, researchers have made considerable progress in analyzing the sequence characteristics of proteins that can undergo phase separation under physiological conditions. The common feature of LLPS proteins is the presence of an intrinsic disordered region (IDR) with multiple interacting motifs. Charge pattern, amino acid composition, and solubility also affect phase separation. We calculated the disorder scores of the proteins by IUPred, the per-residue prion-like scores were obtained from PLAAC, exposure and surface accessibility analysis were performed by NetSurfP, the hydrophobicity was based on the theory of Kyte, J et al., as well as charge from Fauchere et al., and we used StatSEG (https://github.com/jszym/StatSEG) to obtain the low-complexity region scores. In addition, we also considered the polarity of amino acids.
Composition of k-spaced amino acid pairs (CKSAAP)
Similar to Zhao et al., we used the composition ratio of residue pairs of k intervals in the protein sequence fragments in the sequence to establish a mathematical model and extract feature vectors. In other words, if a peptide consists of 20 kinds of amino acids, each amino acid and its next adjacent amino acid form a pair of extracted amino acids, that is, the separation distance between these two amino acids is k = 0 amino acids, then there are 400 possible amino acid pairs (e.g., AA, AC, AD, and so on). According to the probability of these residue pairs appearing in this protein sequence, a 400-dimensional feature vector is generated. With the increase in the k value, although the accuracy and sensitivity of the prediction model increases, the calculation time and cost of the random forest model training also increases notably. In this regard, only the CKSAAP coding with k values equal to 0, 1, 2, and 3 are considered; therefore, the total dimension of the feature vector is 400 × 4 = 1,600.
Position-specific scoring matrix (PSSM)
PSSM is a common feature extraction method in biological sequence analysis, also known as the position weight matrix. This matrix has 20 × M elements, where M is the length of the target sequence. The occurrence frequency of different amino acids at each position in the matrix was calculated, and the details are as follows:

In Eq. 1, the peptides consist of 15 amino acids; P(X1) represents the occurrence frequency of amino acid AA1 at position 1 in the positive group, while N(X1) denotes the occurrence frequency of amino acid AA1 at position 1 in the negative group. Therefore, each peptide can be represented by a position weight amino acid composition vector with dimensions of 30.
Amino acid composition (AAC)
AAC is an elementary feature and describes the frequency of occurrence of each amino acid in the sequence. The dimension of AAC is 20 in this work. Binary encoding profiles (BE) Binary encoding is similar to the binary language of computers [36]. We converted each sequence into a combination of 20-dimensional vectors. For example, if a sequence is ARDCQEHIGNLKMFPSTWYV, then amino acid A corresponds to (10000000000000000000), and amino acid V corresponds to (00000000000000000001). In this work, the vector size is 300.
Option explanation
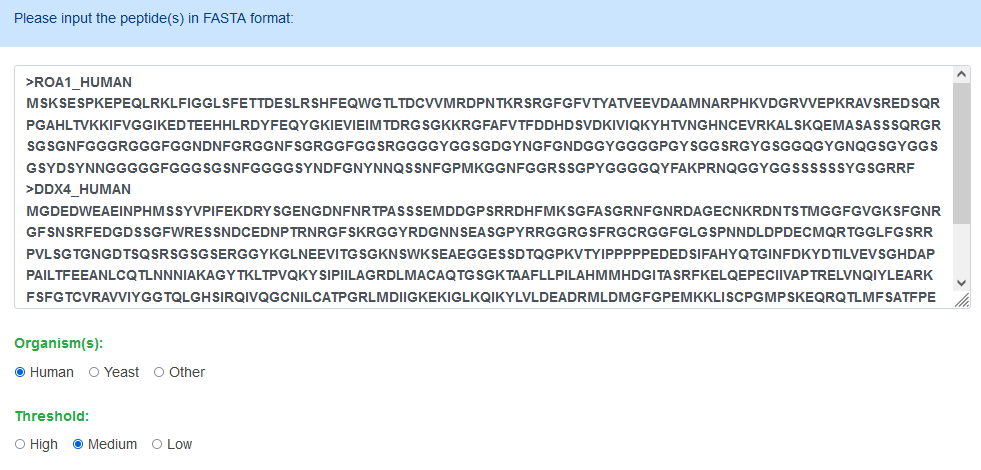
Searching text: Users should input protein sequence in sequence or FASTA format, otherwise will return no results.
Organism: Users can select different organisms to predict, the model performed well in human and yeast. For other organisms, you can use our model to predict the potential regions, but we are not sure about the accuracy.
Threshold: High means a false positive rate at 1%, Medium means a false positive rate at 5% and the Low means a false positive rate at 10%.
